How to use BioMart
Summary
- What is BioMart
- Select a mart database (a type of data)
- Select a mart dataset (a species)
- Filter your mart query (query restriction and input data)
- Select mart attributes (desired output)
- Display and retrieve your query
What is BioMart
BioMart is an easy-to-use web-based tool that allows extraction of data without any programming knowledge or understanding of the underlying database structure. You can navigate through the BioMart web interface using the left panel. Filters and attributes can be selected in the right panel. A summary of your choices is also displayed in the left panel.
1. Select a mart database (a type of data)
First, select a mart database which will correspond to the type of data you are interested in. In Ensembl, you can choose data from one of our four mart databases:
- Ensembl Genes: This mart contains the Ensembl gene set and allows you to retrieve Ensembl genes, transcripts and proteins as well as external references, microarrays, protein domains, structure, sequences, variants (only variants mapped to Ensembl Transcripts) and homology data.
- Ensembl Variation: This mart allows you to retrieve germline and somatic variants as well as germline and somatic structural variants. This mart also contains variants' phenotypes, citations, synomyms, consequences and flanking sequences; you can also retrieve Ensembl genes, transcripts, regulatory and motif features mapped to variants.
- Ensembl Regulation: This mart allows you to retrieve regulatory features, evidence and segments, miRNA target regions, binding motifs and other regulatory regions.
- Vega: This mart contains the Ensembl Vega gene set (manual annotation coming from Havana) and allows you to retrieve Ensembl Vega genes, transcripts and proteins as well as external references, structures, sequences and protein domains
2. Select a mart dataset (a species)
Next, select a mart dataset which correspond to the species you are interested in and want to retrieve data from.

- Ensembl Genes: All the Ensembl species, full list on the species page.
- Ensembl Variation: We only have a subset of our species which have variation data available, full list on the variation species page.
- Ensembl Regulation: We have regulation data available for human, mouse and fruitfly.
- Vega: We have data available for human, mouse, rat, pig and zebrafish.
3. Filter your mart query (query restriction and input data)
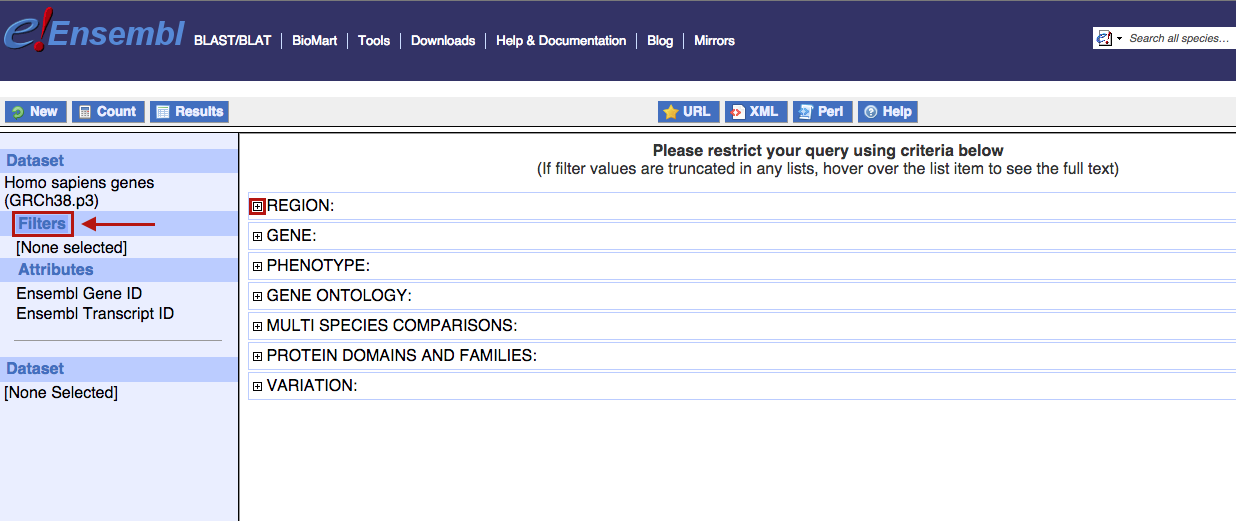
BioMart allows you to restrict your query with information that you know, e.g: input a list of IDs, restrict to a region. You can access the filter page by clicking on the "Filters" button located on the left panel. Filters are organised into different sections, clicking on the "+/-" boxes will expand/collapse a section and display its content.
In the following image, I have expanded the "Region" section by clicking on the "+" box as indicated by the number 1. Then, I have selected "1" in the chromosome list to restrict my query to all the genes located on human chromosome 1. The left panel now displays "Chromosome: 1" as indicated by the number 2. Clicking on the "Count" button as indicated by number 3 will display the number of Ensembl genes that the query will return next to "Dataset" as indicated by the number 4. This mean that we have 5406 Ensembl Genes located on the human chromosome 1.

4. Select mart attributes (desired output)
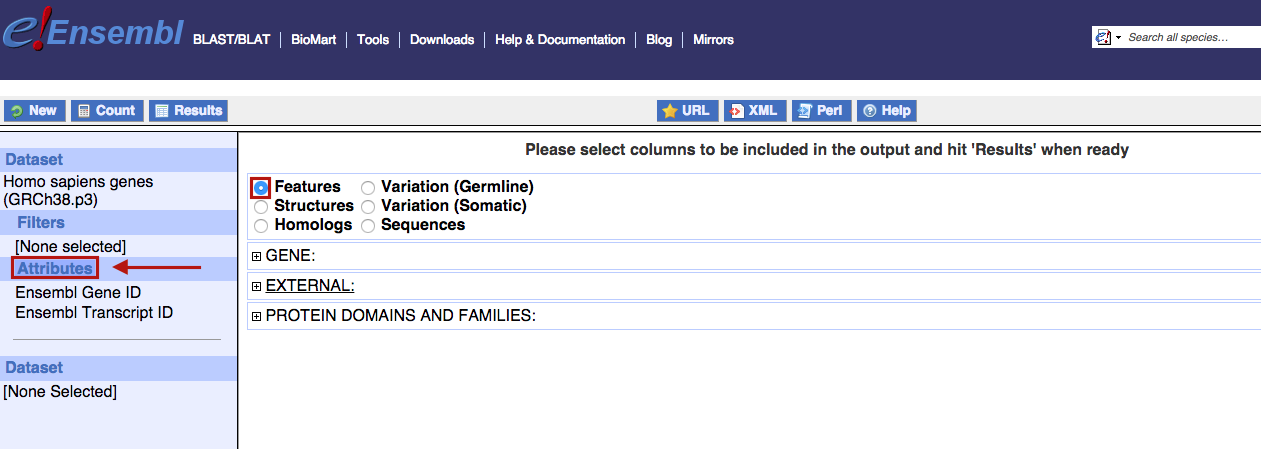
By clicking on the "Attributes" button on the left panel, you will access the mart attribute page. This page allows you to select your desired output; the default output is "Ensembl Gene ID" and "Ensembl Transcript ID" in the Ensembl Genes mart. The attributes are organised in pages that you can access by selecting the radio buttons as displayed by the red box on the image below.
5. Display and retrieve your query
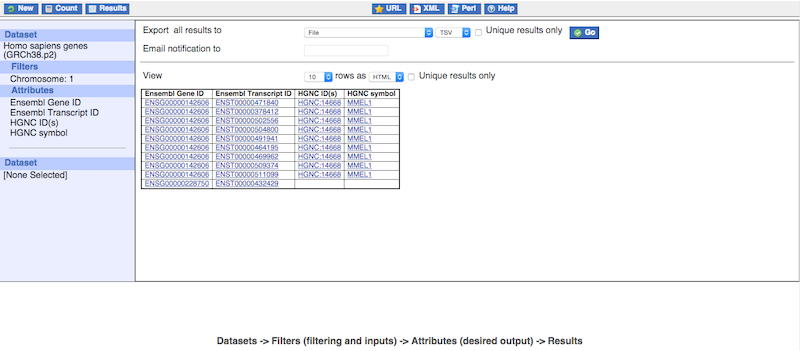
Clicking on the "Results" button will bring you to the mart result page. This page will, by default, show you a preview of the first 10 results of your query in HTML format. The number of results previewed and format can be changed as indicated by number 1 on the image below. You can also automatically remove all the duplicated results from your query by clicking on the "Unique results only" button as indicated by number 2. If you are happy with your query, you can use the "Export all results to" section as indicated by number 3 (select a format and click on the "Go" button) to download your results.

You can also request the results of your query to be sent to you by email. To do this, select "Compressed web file (notify by email)" in the first dropdown in the red box on the image below, then select your desired format, type your email address in the "Email notification to" box and press the "Go" button. BioMart will compile the result of your query in the background and send you a link to the compressed file by email.

BioMart tutorials, multiple dataset query, Perl API, RESTful and Bioconductor R package access documentations can be found on the following page. If you have any questions, you can email Ensembl helpdesk using our web form.
