Customising the species list on your home page
If you are installing a subset of the standard Ensembl species, you can leave the plugins as-is and your home page will be generated automatically.
If, however, you are making major changes to the list of species found in Ensembl, you may want to make changes to how those species are listed.
For a site with few species, it is usually sufficient to provide a list of static links, or
a simple list can be auto-generated using the information stored in SpeciesDefs. See
EnsEMBL::Web::Document::HTML::SpeciesList for sample code.
If your site has a lot of species, you may wish to automatically group these by taxa, in a similar way to Ensembl.
Organising your species
To automatically group species by taxa, you need taxonomy data in the 'meta' table of your core database. The standard format is for the taxa to be listed in ascending order when ordered by meta_id, e.g.
mysql> select * from meta where meta_key = 'species.classification' order by meta_id; +---------+------------------------+------------------+ | meta_id | meta_key | meta_value | +---------+------------------------+------------------+ | 1039 | species.classification | sapiens | | 1040 | species.classification | Homo | | 1041 | species.classification | Hominidae | | 1042 | species.classification | Catarrhini | | 1043 | species.classification | Haplorrhini | | 1044 | species.classification | Primates | | 1045 | species.classification | Euarchontoglires | | 1046 | species.classification | Eutheria | | 1047 | species.classification | Mammalia | | 1048 | species.classification | Euteleostomi | | 1049 | species.classification | Vertebrata | | 1050 | species.classification | Craniata | | 1051 | species.classification | Chordata | | 1052 | species.classification | Metazoa | | 1053 | species.classification | Eukaryota | +---------+------------------------+------------------+
When provided with a list of taxa, SpeciesDefs will compare the two in order to classify your species into groups.
The next step is therefore to identify the taxa that correspond to the groups you want. In Ensembl, we have many different mammal species, some other vertebrates, and a few non-vertebrates. We have thus divided our species as follows:
- Primates
- Other Euarchontoglires (rodents and so forth)
- Laurasiatheria
- Afrotheria
- Xenarthra
- Marsupials and Monotremes
- Birds and Reptiles
- Amphibians
- Fish
- Other chordates*
- Other eukaryotes
* Although this is a very broad set of phyla, they have few representative genomes, having been sequenced primarily for comparative purposes, so we list them together.
Once you have decided how to group your species, you need to sketch a taxonomic tree that includes the identifying nodes. In the diagram below, the highlighted taxa are the ones that will be needed to separate species into the required groups.
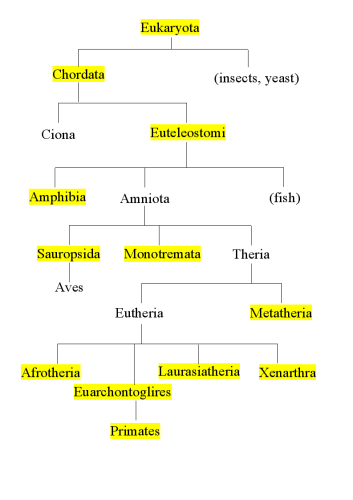
Finally you will need to set up your [plugin]/conf/ini-files/DEFAULTS.ini to contain the groups you wish to select and the corresponding labels. The order of the groups must follow your taxonomic tree from bottom to top:
TAXON_ORDER = [Primates Euarchontoglires Laurasiatheria Afrotheria Xenarthra Metatheria Monotremata Sauropsida Amphibia Euteleostomi Chordata Eukaryota]
If you want any of these taxa to be grouped together, or to use labels other than their scientific name, you also need a hash of labels:
[TAXON_LABEL] Euarchontoglires = Rodents etc. Metatheria = Marsupials & Monotremes Monotremata = Marsupials & Monotremes Sauropsida = Birds & Reptiles Amphibia = Amphibians Euteleostomi = Fish Chordata = Other chordates Eukaryota = Other eukaryotes
Note that by setting the same group label for different taxa, we can combine taxonomic groups that are in different branches of the taxonomic tree
Caveats
A few things that could potentially cause problems are:
- Each species ini file must contain the block:
[TAXON_LABEL]
# Accept defaults - If you are upgrading from an earlier version of Ensembl, please check that each species ini file has the parameter ENSEMBL_GROUP deleted, as it is now generated automatically.
- If you are upgrading from Ensembl 48 to 49, the taxon parameters in DEFAULTS.ini have changed slightly to take into account Perl's inability to keep the order of elements in a hash.
