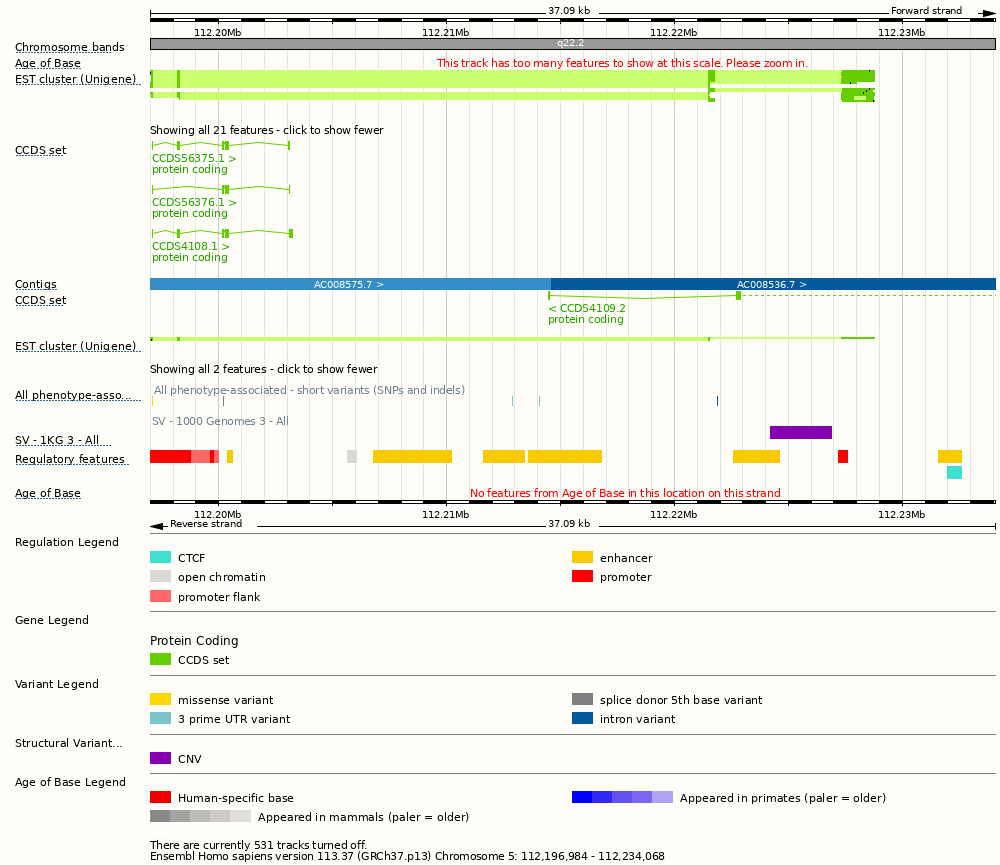
[["rect",[150,38,996,50],{"title":"Band: q22.2","href":"/Homo_sapiens/ZMenu/Location/View?config=contigviewbottom;db=core;r=5:111500001-113100000;track=chr_band_core"}],["rect",[15,38,112,52],{"klass":["label","nwd458j4"],"alt":"Human (Homo sapiens)"}],["rect",[15,54,82,68],{"alt":"Human (Homo sapiens)","klass":["label","lwtKMlEH"]}],["rect",[708,100,715,102],{"title":"UniGene Eca.1570","href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Eca.1570;ln=unigene;r=5:112221479-112221763;track=dna_align_core_unigene","klass":["group"]}],["rect",[842,100,875,102],{"title":"UniGene Nvi.10076","klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Nvi.10076;ln=unigene;r=5:112227379-112228743;track=dna_align_core_unigene"}],["rect",[841,98,874,100],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Spu.24257;ln=unigene;r=5:112227349-112227795;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Spu.24257"}],["rect",[841,96,874,98],{"title":"UniGene Dm.4433","klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Dm.4433;ln=unigene;r=5:112227340-112227789;track=dna_align_core_unigene"}],["rect",[841,94,875,96],{"title":"UniGene Dr.151349","href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Dr.151349;ln=unigene;r=5:112227340-112227801;track=dna_align_core_unigene","klass":["group"]}],["rect",[841,92,875,94],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=At.51551;ln=unigene;r=5:112227337-112227984;track=dna_align_core_unigene","title":"UniGene At.51551"}],["rect",[708,90,873,92],{"title":"UniGene Mm.469937","klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Mm.469937;ln=unigene;r=5:112221479-112221763;track=dna_align_core_unigene"}],["rect",[708,88,875,90],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Cfa.242;ln=unigene;r=5:112221479-112221763;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Cfa.242"}],["rect",[179,98,715,100],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Tgu.2576;ln=unigene;r=5:112198266-112198274;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Tgu.2576"}],["rect",[151,96,710,98],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Pmn.6066;ln=unigene;r=5:112197071-112197112;track=dna_align_core_unigene","title":"UniGene Pmn.6066"}],["rect",[151,94,709,96],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Oar.4996;ln=unigene;r=5:112197029-112197112;track=dna_align_core_unigene","title":"UniGene Oar.4996"}],["rect",[150,92,710,94],{"title":"UniGene Str.11353","href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Str.11353;ln=unigene;r=5:112197026-112197112;track=dna_align_core_unigene","klass":["group"]}],["rect",[150,86,710,88],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Pab.13620;ln=unigene;r=5:112197017-112197112;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Pab.13620"}],["rect",[150,84,710,86],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Tvu.2485;ln=unigene;r=5:112197020-112197112;track=dna_align_core_unigene","title":"UniGene Tvu.2485"}],["rect",[150,82,857,84],{"title":"UniGene Gac.8426","klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Gac.8426;ln=unigene;r=5:112196996-112197112;track=dna_align_core_unigene"}],["rect",[151,80,875,82],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Ssc.75017;ln=unigene;r=5:112197068-112197112;track=dna_align_core_unigene","title":"UniGene Ssc.75017"}],["rect",[151,78,875,80],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Rn.116072;ln=unigene;r=5:112197068-112197112;track=dna_align_core_unigene","title":"UniGene Rn.116072"}],["rect",[151,76,875,78],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Gga.43958;ln=unigene;r=5:112197065-112197112;track=dna_align_core_unigene","title":"UniGene Gga.43958"}],["rect",[150,74,875,76],{"title":"UniGene Hs.637001","klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Hs.637001;ln=unigene;r=5:112197008-112197112;track=dna_align_core_unigene"}],["rect",[150,72,875,74],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Mmu.11319;ln=unigene;r=5:112196984-112197112;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Mmu.11319"}],["rect",[150,70,875,72],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Bt.49410;ln=unigene;r=5:112196981-112197112;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Bt.49410"}],["rect",[150,126,996,135],{"href":"/Homo_sapiens/ZMenu/Location/ExpandTrack?config=contigviewbottom;count=21;db=core;default=40;goto=View;r=5:112196984-112234068;track=dna_align_core_unigene"}],["rect",[15,70,142,84],{"klass":["label","danSZVnJ"],"alt":"Human (Homo sapiens)"}],["rect",[152,141,290,150],{"title":"Transcript: CCDS56375.1; Gene: CCDS56375.1; Location: 5:112197074-112203100","klass":["group"],"href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS56375.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS56375.1;track=transcript_otherfeatures_ccds_import"}],["rect",[152,149,331,163],{"title":"Transcript: CCDS56375.1; Gene: CCDS56375.1; Location: 5:112197074-112203100","href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS56375.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS56375.1;track=transcript_otherfeatures_ccds_import"}],["rect",[152,141,290,194],{"href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS56376.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS56376.1;track=transcript_otherfeatures_ccds_import","klass":["group"],"title":"Transcript: CCDS56376.1; Gene: CCDS56376.1; Location: 5:112197074-112203112"}],["rect",[152,193,331,207],{"href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS56376.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS56376.1;track=transcript_otherfeatures_ccds_import","title":"Transcript: CCDS56376.1; Gene: CCDS56376.1; Location: 5:112197074-112203112"}],["rect",[152,141,293,238],{"title":"Transcript: CCDS4108.1; Gene: CCDS4108.1; Location: 5:112197074-112203234","href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS4108.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS4108.1;track=transcript_otherfeatures_ccds_import","klass":["group"]}],["rect",[152,237,325,251],{"title":"Transcript: CCDS4108.1; Gene: CCDS4108.1; Location: 5:112197074-112203234","href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS4108.1;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS4108.1;track=transcript_otherfeatures_ccds_import"}],["rect",[15,141,64,155],{"klass":["label","jz1wZ5oP"],"alt":"Human (Homo sapiens)"}],["rect",[150,278,552,290],{"href":"/Homo_sapiens/ZMenu/Location/Contig?config=contigviewbottom;db=core;r=5:112196984-112214602;region=AC008575.7;track=contig","title":"AC008575.7"}],["rect",[551,278,996,290],{"title":"AC008536.7","href":"/Homo_sapiens/ZMenu/Location/Contig?config=contigviewbottom;db=core;r=5:112214603-112234068;region=AC008536.7;track=contig"}],["rect",[15,275,58,289],{"klass":["label","8gJPI1sE"],"alt":"Human (Homo sapiens)"}],["rect",[548,291,996,300],{"title":"Transcript: CCDS4109.2; Gene: CCDS4109.2; Location: 5:112214483-112257887","href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS4109.2;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS4109.2;track=transcript_otherfeatures_ccds_import","klass":["group"]}],["rect",[548,299,721,313],{"title":"Transcript: CCDS4109.2; Gene: CCDS4109.2; Location: 5:112214483-112257887","href":"/Homo_sapiens/ZMenu/Transcript/CCDS?calling_sp=Homo_sapiens;config=contigviewbottom;db=otherfeatures;g=CCDS4109.2;r=5:112196984-112234068;real_r=5:112196984-112234068;t=CCDS4109.2;track=transcript_otherfeatures_ccds_import"}],["rect",[15,291,64,305],{"klass":["label","pthFq6Zg"],"alt":"Human (Homo sapiens)"}],["rect",[150,337,875,339],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Hs.640499;ln=unigene;r=5:112197026-112197112;track=dna_align_core_unigene","title":"UniGene Hs.640499"}],["rect",[150,339,710,341],{"href":"/Homo_sapiens/ZMenu/Location/Genome?config=contigviewbottom;db=core;ftype=DnaAlignFeature;id=Pan.3461;ln=unigene;r=5:112196987-112197112;track=dna_align_core_unigene","klass":["group"],"title":"UniGene Pan.3461"}],["rect",[150,365,996,374],{"href":"/Homo_sapiens/ZMenu/Location/ExpandTrack?config=contigviewbottom;count=2;db=core;default=40;goto=View;r=5:112196984-112234068;track=dna_align_core_unigene"}],["rect",[15,337,142,351],{"klass":["label","TG3vs04A"],"alt":"Human (Homo sapiens)"}],["rect",[152,396,153,406],{"href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-11-rs199905553-3-vdb-9-variation-2-vf-9-329528918-","title":"Variation: rs199905553; Location: 112197084; Consequence: missense_variant; Ambiguity code: B"}],["rect",[152,396,153,406],{"href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-11-rs367813945-3-vdb-9-variation-2-vf-9-329659880-","title":"Variation: rs367813945; Location: 112197104; Consequence: missense_variant; Ambiguity code: V"}],["rect",[223,396,224,406],{"title":"Variation: rs1322282571; Location: 112200230; Consequence: splice_donor_5th_base_variant; Ambiguity code: R","href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-12-rs1322282571-3-vdb-9-variation-2-vf-9-356880689-"}],["rect",[512,396,513,406],{"href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-11-rs114461222-3-vdb-9-variation-2-vf-9-327833823-","title":"Variation: rs114461222; Location: 112212914; Consequence: 3_prime_UTR_variant; Ambiguity code: S"}],["rect",[539,396,540,406],{"title":"Variation: rs2546115; Location: 112214093; Consequence: 3_prime_UTR_variant; Ambiguity code: H","href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-9-rs2546115-3-vdb-9-variation-2-vf-9-326677502-"}],["rect",[717,396,718,406],{"href":"#:@:6-config-16-contigviewbottom-8-snp_fake-1-1-7-species-12-Homo_sapiens-5-track-25-variation_set_ph_variants-4-type-9-Variation-1-v-8-rs818427-3-vdb-9-variation-2-vf-9-326606186-","title":"Variation: rs818427; Location: 112221869; Consequence: intron_variant; Ambiguity code: Y"}],["rect",[15,386,142,400],{"alt":"Human (Homo sapiens)","klass":["label","kIZAdZID"]}],["rect",[770,426,832,439],{"href":"/Homo_sapiens/ZMenu/StructuralVariation/View?config=contigviewbottom;db=core;r=5:112196984-112234068;sv=esv3606302;svf=45696332;track=sv_set_1kg_3;vdb=variation","klass":["group"],"title":"Structural variation: esv3606302; Source: DGVa; Location: Chr 5:112224227-112263971"}],["rect",[15,426,112,445],{"alt":"Human (Homo sapiens)","klass":["label","TyBIDgCF"]}],["rect",[150,450,201,463],{"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00000185353;track=regulatory_build","klass":["group"]}],["rect",[200,450,219,463],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00002015772;track=regulatory_build"}],["rect",[227,450,233,463],{"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00002015773;track=regulatory_build","klass":["group"]}],["rect",[347,450,357,463],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00002015774;track=regulatory_build"}],["rect",[373,450,452,463],{"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001697152;track=regulatory_build","klass":["group"]}],["rect",[483,450,525,463],{"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001697154;track=regulatory_build","klass":["group"]}],["rect",[528,450,602,463],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001337080;track=regulatory_build"}],["rect",[733,450,780,463],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001337082;track=regulatory_build"}],["rect",[838,450,848,463],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00002015775;track=regulatory_build"}],["rect",[938,450,962,463],{"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001337084;track=regulatory_build","klass":["group"]}],["rect",[947,450,962,479],{"klass":["group"],"href":"/Homo_sapiens/ZMenu/Regulation/View?config=contigviewbottom;db=core;fdb=funcgen;r=5:112196984-112234068;rf=ENSR00001337085;track=regulatory_build"}],["rect",[15,450,130,464],{"alt":"Human (Homo sapiens)","klass":["label","CJxqC0Iq"]}],["rect",[15,484,82,498],{"alt":"Human (Homo sapiens)","klass":["label","XHgrMoFJ"]}],["rect",[15,533,118,547],{"klass":["label","hToKFeVB"],"alt":"Human (Homo sapiens)"}],["rect",[15,611,82,625],{"klass":["label","A8yYEIKC"],"alt":"Human (Homo sapiens)"}],["rect",[15,675,100,689],{"klass":["label","tPp0MH4z"],"alt":"Human (Homo sapiens)"}],["rect",[15,734,142,748],{"alt":"Human (Homo sapiens)","klass":["label","Slo34YPu"]}],["rect",[15,773,124,787],{"klass":["label","CGs3J70J"],"alt":"Human (Homo sapiens)"}],["rect",[150,5,996,532],{"alt":"Click and drag to select a region","href":"#drag|1|3|Homo_sapiens|5|112196984|112234068|1","klass":["drag"]}]]



 Collapsed
Collapsed